What Every Veterinary Oncologist Needs to Know About Cancer Genomics
Genomics is fast becoming an essential part of veterinary oncology. It touches upon all aspects of practice, from diagnostics to therapeutics. Genetic information can be incredibly useful in terms of making a more accurate diagnosis or making a more precise prognosis, or even determining what therapy to use or what order to use different therapies.
Dr. Shaying Zhao is a pioneer in the field of comparative oncology. She is a scientist, professor, and researcher. Dr. Zhao was the first to propose and use novel dog-human comparison strategy for cancer driver-passenger discrimination, a central aim of cancer research. Since 2005, her research has focused on dog-human comparative genomics and oncology.
Her work looks at and decodes the canine cancer genomic landscape and is used to figure out what is similar to the human landscape, what's not, and where can we bring in interventions.
This webinar discusses the value of looking at genomes comparatively, the value of big data and the value it can bring to both sides of the leash.
What are your academic interests and why did you choose to study genomics.
I earned a PhD in biochemistry and worked at TIGR (The Institute for Genomics Research now called JCVI), the institute founded by Craig Venter (the first to sequence the human genome). That's where I learned genomics and bioinformatics while part of a team sequencing the human, mouse, rat, and cow genomes. After seven years, in 2005, I came to the University of Georgia to start my own lab in the cancer center. In my lab we work on something super exciting and important - Cancer.
The cancer field is really competitive. So I had to find my own niche, that's why I turned to the dog and focused on comparative genomics and oncology. That's why I initially built my lab, it has been 15 years. My team is totally devoted to comparative genomics oncology between dogs and humans.
In terms of the canine or comparative genomics, we talked about big data and bioinformatics. Can you tell us about what that means to you, and why you're excited to work in this field with “big data”, with the comparative aspects of dog and human genomics?
We focus on big data but on a really particular part of big data, on genomics. We analyzed hundreds of canine genomes sequences by the FidoCure® team and others. There are hundreds of tumor genomes being sequenced for the dog. We use that data and then we also bring the human data which is, tens of thousands of genomes that have been sequenced. It's just a tremendous amount of data, really exciting.
My team has the power to analyze tens of thousands of genomes and extract meaningful information from them. We use software tools, which were often developed for human cancer, and it has been working fantastically for us.
The data is really exciting, especially for the canine. The human tumor genome has been sequenced, probably the past year, so now the data is kind of saturated. For the dog it's different, because everyday, we have genomes deposited into the database. The data is growing exponentially.
This is a really exciting time to be in comparative genomics and oncology, particularly on the dog side.
When you talk about the thousands upon thousands of genomes that you have access to, or that you can interrogate really on a daily basis because of your ability to look at multiple different data sets in terms of canine genomics and compare them to human genomics, I can imagine that there aren't very many people or groups who have access to the amount of data, both human and canine that your group has access to.
The reason we can do that is because the University of Georgia has really powerful computing resources. I'm also fortunate to have a team of graduate students focused on the large data sets. And then the other thing that I really appreciate is the experience I gained from TIGR, the institute that's funded by Craig Venter. It's a really different science that totally opened up my mind.
We are looking for big data, looking for big pictures. And one genome is not enough, hundreds of genomes are not enough. We are looking for thousands of genomes, tens of thousands of genomes, and when you have this type of data, you are looking at a really broad picture, and then the exciting information will come out.
That's fantastic. What were some of the key lessons that you learned from the genomics institute -TIGR- that you brought to the canine world of genomics?
One thing TIGR is really famous for is high quality data. The data quality is really the core. In big data just like any other science, we need high quality data, particularly for cancer.
The genomics, the bioinformatics, are really only two aspects. There is so much more we need: pathology; tumor biology; histology; treatment course; and treatment response. We need comprehensive data sets. And that's really what- once we bring everything together- really makes data useful.
The other thing is that TIGR always strives for high quality data, like the human genome sequencing, TIGR did a lot of microbial genome sequencing plans. The data from TIGR is always high quality, and that's what I learned from Craig and the others at TIGR. For the canine field, what we really need to achieve is generating as accurate data and as comprehensive data as the human cancer field.
Canine cancer is just fantastic. We have let's say 80 million canine pets in the US. Those dogs get cancer naturally, which is really unfortunate. This is a really underutilized resource. What we can do with comparative genomics--studying human and canine cancer--where there's lots of information that can be extracted from that comparison. We get much more information than just studying human cancer alone.
In my opinion, comparative oncology is the most underutilized toolbox in cancer research, and I am perfectly in agreement with you that it's an incredibly powerful tool that should be used more and more and more. So yes, please share whatever data you want to show us.
My students are analyzing the FidoCure® data, including the mutation landscape for the gene encoded TP53. It's a tumor suppressor gene. This gene is the most frequently mutated gene in both human and dog. If you're looking for the mutation landscape, there's tens of thousands of human cancer data compiled. And for the dog, this FidoCure® data, there's more than 700 cases.
If you analyze the dog and human together, you see this incredible homology between dogs and humans. Because it's a tumor suppressor gene, when this gene mutated or lost its function, then the cancer will develop. In both the human and the dog these mutations are scattered, literally matching, and sometimes you see the exact same mutations.

In the TP53 tumor suppressor gene, we see a tremendous homology between dog and human. I don't think there's any effective drug targeting these mutations, but if there was a drug that was developed in the future, targeting TP53 mutations, it would be great.
With the current system one of the problems in drug discovery is that it would really take 10 years and one billion dollars to bring a drug to the market. The reasons for the high cost and long time is because many of the preclinical drugs fail in the stage three clinical trials. Most people are using the mouse model or human cell lines to do the preclinical drug discovery. Those really don't match the experience of these drugs in people very well. So even though they’re really successful in the preclinical research, when you try these drugs in people, they just simply fail. I think over 90% of the preclinical drugs just fail at stage three human clinical trials.
The goal is that, because the dog is so similar to the human and there's no FDA regulation on the dog, we actually can do effective clinical trials. Because dogs and humans are so similar, in terms of their cancer biology, in terms of the size and the dose response, we can try a drug in the dog. If it's working for the dog, then offer that trial to humans; it will increase the success rate of the drug discovery treatment. I think it will significantly decrease the overall cost, and decrease the time for anticancer drug development. So that's the homology side benefit to dogs and humans.
The other example of the FidoCure® dataset, involves another gene, called PIK3CA. When mutation happens in TP53, the gene just loses its function. That's a coded tumor suppressor. When the mutation happens in PIK3CA, it is essentially activating the gene so the gene becomes over activated, which also leads to cancer development.
If you're looking at the human data, this is compiled from tens of thousands of human data, and immediately you see two mutation hotspots. We examine this same thing in the dog. The mutation hotspots were completely identically shared between the dog and the human. But humans have another mutation hotspot, but in the dog, it just disappeared. So the question is why, right?
We see homology, which is great but we also see a difference. Right now, these two hotspots mutation has been reported for quite a few years and people are starting to look at it, but we still don't know its function. What were the hotspots doing in terms of biology or cancer biology? The dog can help answer those questions, because we have a negative control, we have a positive control, and we can study both the homology and the difference.
Once there are FDA approved drugs targeting those mutations, once we understand the mutations, we can use these drugs to treat the canine mammary tumors. That’s how the data produced by FidoCure® is great.
Thank you so much for sharing the data. Just to step back just a little. One of your lab's key areas of interest is looking at driver mutations, as it relates to certain cancers, so driver mutations and passenger mutations. Can you go over what your lab's interests are in this particular area and how you correlate driver mutations, passenger mutations when you're looking at dogs versus humans?
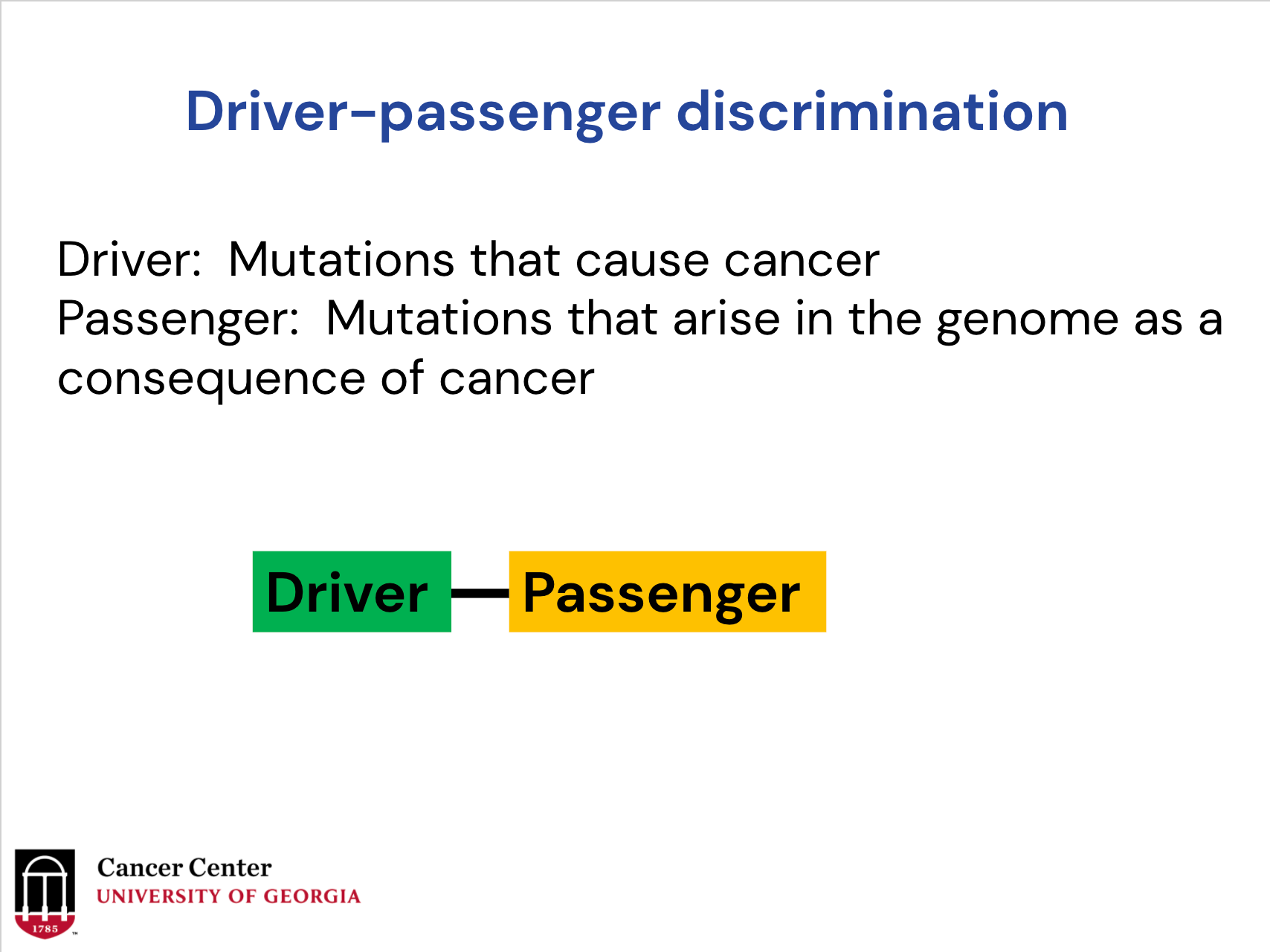
Driver mutations are actually the mutations that cause the cancer. They contribute to the cancer initiation and progression. We also have the passenger mutations, mutations that do not drive the cancer.
When you sequence the tumor genome, you will find hundreds to thousands of mutations. Only a small portion of the vast majority of them arise in the genome because of the cancer. Those mutations, we call the passenger.
If you're a drug company, you want to target the driver mutation not the passenger mutations. So that's why driver/passenger discrimination has always been a central aim of cancer research.
Let me show you how we achieve driver/passenger discrimination.

This is data from mostly FidoCure® and our data. We have not published this paper yet but we combined both data sets together, and this actually shows vast mutations. The reason this is exciting is that actually, this is an actionable mutation. And then if you're looking for the human compared to the dog, they actually share exactly the same mutation hotspots, and this is just amazing data. When you do this comparative, I'm just so excited at this type of comparative genomics, oncology data because to me, this is the most rewarding seeing these types of results. Really showing how important the dog is.
As excited as you are, Dr. Zhao, we are equally as excited to collaborate and work with you and your team. It's just been fantastic. If you want to go over the driver/passenger discrimination, I think that would be really fantastic for our audience.
Driver/passenger discrimination is actually a central aim of cancer research. Again, the driver is a mutation that causes cancer, the passengers are mutations that arise in the genome as a consequence of cancer.
This has been a central aim of cancer research. Many approaches have been developed to distinguish the drivers from passengers. Most labs use the rate of recurrence of the mutations to distinguish drivers from passenger mutations. They only study human cancer sequences. Once you sequence tens of thousands of cancers and look for the mutations which occur with the cancers, those cases most likely will be drivers.
The rare mutations, will most likely be classified as passenger mutations. That approach works for some cases, but for some cases, it doesn't work. In the genome, you have two gene mutations, one's a driver, one's a passenger. Some mutation, it actually causes the cancer. Another gene mutated, by itself, it doesn't cause cancer.
We not only see mutations in single base pairs, but in the cancer genome, we also see large scale changes, like a large chunk of DNA just getting lost, or a large chunk of DNA just getting amplified. Normally, we only have two copies of a gene, but in some cancer genomes, we can have multiple copies of the same gene, like eight or 10 copies.
If two genes in the genome are actually neighbors, if you're only looking for the recurrence rate of these genes in tumors, for instance---in how many tumors these genes are amplified or lost, then because they are neighbors--they may get amplified or lost at the same frequency even if one is a driver mutation and one is a passenger mutation. Looking at frequency alone, you cannot distinguish what's a driver or what's a passenger mutation.
When I built my lab, I found this a very interesting problem. We actually take a really different approach than the traditional labs, and that approach is focused on the dog.
Each one of us has 22 autosomal genes, females have two Xs and the males have X and Y. So is actually how the human genome looks like. This is dog genomes. What I was showing over here is we give each canine chromosome a distinct color and then present the human genome using the canine chromosomes. You can chop the dog chromosome into 400 pieces, reshuffle them and then you form the human genomes.

This is great because we can identify genes which are amplified in human cancers, and then we can do the same thing with the dog. First thing, for those tumors, we have to make sure that they're molecularly identical, and we're doing this for colorectal cancer. We show that a canine colon cancer just like human cancer, they are really similar. They share the same tumor progression pathways. And then they share the part, they commonly amplify the genes that will be drivers. So that's the first driver we identified.
We cannot only identify those drivers. We actually can distinguish the drivers from passengers. I already told you two neighbor genes in the human genome, which were amplified together. So just by studying human cancer, we don't know which one is the driver, which one is the passenger because they happened at equal frequency. But in the dog, because the genomic rearrangements, they can stay really far away. They are no longer neighbors. And they can even be located on different chromosomes.
I'll give you a real example. Those are two gene neighbors, located on human chromosome one. They are amplified together in the human colon cancer. But in the dog, they are separate. Those In the dog, one is located on chromosome five, another one is located on chromosome six. So this is a real driver gene. If this one lost its function or becomes abnormal, then the DNA damage will accumulate in the genome and that essentially leads to cancer.

But in the dog, only one gene was amplified. We say that we know this is a driver gene, and both of them, we see this amplified also. But this other one, we classify as a passenger, and no study reporting that it is contributing to the cancer, and this one is located in the chromosome five, so whatever this driver is doing, it really doesn't affect this gene because it's so far away. They're no longer neighbors.
We take this approach and apply to the entire genome and showing it is really simple but powerful. So that's how we take advantage of the dog/human comparison of the cancer research.
That is fascinating, Dr. Zhao. In other words, what your lab has done over the years is utilize the power of comparative genomics, comparing dog genome and human genome, dog cancer genome, human cancer genome, to discriminate the driver mutations in human cancers.
Yes. That's what they do.
That's so fascinating because certainly from our perspective--and I know we're a little bit biased ---we're also incredibly concerned about what the driver mutations are in canine cancers.
Yes, we can do the other way around.
So utilizing “both sides of the leash” and information flowing in both directions is just fascinating.
Looking at osteosarcoma, oral melanomas and hemangiosarcoma, there's lots of happening in the dog. So we can use it the other way around. As long as we are showing they are molecularly matched, right? We are comparing apples to apples, not an apple to orange, then we can use this really simple powerful approach, looking for the changes.
This method also works for epigenetic changes and I think epigenetic changes are also really important for canine cancer. Not just genetic changes, perhaps gene expression changes too.
Is your lab also working on the epigenetic changes in comparison?
We want to. We're still really much focused on the genetic changes, but in the future, I think the epigenetic changes probably perhaps even more important to the canine cancer than the human adult cancer. We know epigenetics is very important in human pediatric cancer. Canine cancer is sort of like between adult and pediatric cancer. Sometimes it actually matches the pediatric cancer very well. That's one of the directions we really want to get into.
Fantastic. How important do you think genomics will be to veterinary oncology given that you really have looked at canine cancer genomics for quite some time.
Extremely important. Because this one leads to more precise medicine. The precision medicine, if a dog gets a tumor, you sequence its genome and then you identify what gene was mutated and what drugs are developed for the human, and you can directly apply that to the canine patient. That's what FidoCure® is doing and that's really fantastic.
The other thing is that once you integrate the dog and the human together, I think this is not only great ... you can look for the treatment and response, I think those data are not only important for the canine but incredibly important for the human field also. Because it's in another species, you learn a lot.
Again, from our perspective, this collaboration with you and your group has been just incredibly rewarding. Now that you have the audience of other veterinary oncologists, what would you tell other veterinarians, as well as other researchers like yourself, about how they can work more closely together to benefit the field of oncology in general?
Genomics and bioinformatics is really just analyzing the data, right? Whatever we do, the results are only as accurate as the data itself. We need genomics and bioinformatics, not just one field, in order to really get to our goals of treating all the cancers, being able to cure all the cancers in the dog or in the human. Oncology is very important. Pathology is very important, Histology is very important. Immunology is very important. Everybody has to work together.
The way I think of this is really great for us, for our students like Josh Watson and Yuan Feng to work with FidoCure®, and they really enjoy the process. Not only looking for the data, the amazing data FidoCure® has been generating, but in the process, they tell me that they learned a lot. That's just one example. The way that I think, everybody needs to be collaborating with each other. I'm basically a scientist. I really don't have the expertise or experience really to understand the oncology side. And I don't have access to the patient.
We are collaborating with you guys, but we're also collaborating with other oncologists- such as those at UGA and other places, to get the tumors. We also collaborate with pathologists and they tell us the particular subtype. The oncologists tell us this is responding to this treatment, but not responding to the other treatment etc. So I think talking to each other, collaborating with each other, that's the most important.
The second thing is that I think we need the infrastructure. Again, the canine tumor is such a fantastic resource, but is significantly underutilized. One of the reasons is just that we lack the infrastructure. There's lots of hospitals, they do surgery each day. They just throw the tumor away, right? As a medical waste. It’s a shame because we have a tumor bank that archives those tumors. We have a database putting the patient information, any treatment, any history into the database. Just like a human, we can sequence this tumor. We could study those tumors. That's the infrastructure. I think a world tissue bank and database should be established.
I have graduate students in my lab. I think training the next generation scientists is very important, actually a new graduate student that just started in my lab. She is a PhD student, and she will be doing genomic research but also trained as a veterinarian. When those people want to become independent investigators, this will move them forward. So that's what I'm thinking of.
I appreciate it. Certainly from our perspective, and my personal perspective, the collaborative nature, I would say of many veterinary oncologists is quite high. We love collaborating with people. I think sharing data is part of our DNA. So when we find a partner such as yourself, who truly understands the value of comparative oncology, as it relates to both veterinary medicine, as well as human medicine, it's just incredibly heartwarming.
Thank you for your open mindedness, for your foresight in understanding the value of comparative oncology, and comparative genomics to both people and pets. It really is wonderful.
Q&A
It strikes me that dogs have much more diversity than humans. Has there been any work done by your lab or others comparing dogs to humans, by breed, sex, or other parameters? In other words, are there dog breeds that are better for comparative work than others?
Yes, I can share a story. One of the things I didn't mention about dogs is the breed. There are greater than 300 pure breeds. That's really unique to the dog. This is an advantage to dogs over humans.
The advantage is actually looking for cancer susceptibility genes, right? Using the golden retriever as an example, they are really prone to develop hemangiosarcoma, osteosarcoma, and lymphoma. Golden retriever breeds, in terms of the pure breeds, their genomes are much closer to each other than for example to a boxer.
Why is the golden retriever so prone to osteosarcoma development, but not as prone to others, for instance mammary tumor. They still have mammary tumors but a lot of mammary tumors also come in smaller breeds. We can sequence their genome and look for mutations. So those are the ones that have a breed specific mutation.
And then after that, we associate those mutations with the disease. We identify significant associations, and that's why we can actually identify the cancer predispositions. Like BRCA1, BRCA2, right? Like in humans, some families have a long history of developing breast cancer and one of the reasons is because they have the mutations in their family but not shared by the general population, so they develop breast cancer at a young age and multiple members of them develop breast cancer.
We can use the dog, the pure breeds to discover more mutations like BRCA1, BRCA2 mutations. If they carry that particular mutation, then the chance they develop cancer is higher.
Another example is carcinoma. I have a small grant funded by a canine club for cocker spaniels. They want to know what gene was responsible for that. If you compare an English cocker spaniel to a non-English cocker spaniel, English cocker spaniel dog has a seven times higher chance to develop carcinoma. I got tumors from owners all over the US. I really want to thank them for supporting the research. We just got a tumor and brought samples in from Maryland. We want to sequence those genomes, not only just the tumor genome but the normal genomes, and really compare English cocker spaniel dogs to other dogs and find out what's different. What makes the English cocker spaniel dog so prone to carcinoma development? We are really devoted to that research. Not just the human, but the dog.

In terms of the answer to the question, I think one of the things is that we see different types of canine tumors, I think histologically, you saw even more subtypes than the human, and there's one reason for that. I might be wrong, but this is my hypothesis. I think canine cancer is really a development cancer, like pediatric cancer, because when the dog gets cancer, they're probably 10 years old or older. But when a human gets cancer, most people are 60, 70 years old.
Let's just use mammary cancer as an example. Breast cancer. We know there's many subtypes in human breast cancer. You have triple negative, or you have the estrogen receptor positive. Those types of tumors. Looking at the dog mammary tumor, there are even more histological subtypes than the human. I think one reason is because of stem cells. When the dog gets the tumors, their mammary glands probably still have stem cells. This is one of the reasons I'm mentioning epigenetic. Because making stem cells become like cancer cells is really easy. You don't need a genetic change, just an epigenetic change is enough/
That's why I think in cancer, there's so much differentiation. This is one of the reasons, canine cancer has many subtypes. That's my hypothesis. I don't have any data for that.
Building off of that, we have another question/comment about comparing pediatric cancers in people to pediatric cancers in dogs. In other words, is there value in looking at pediatric cancers in people, to cancers in dogs that occur in dogs that are young?
Yes, we compare the age between a dog and the human. If we see a tumor in four years or younger dogs, that's probably sort of equivalent to the pediatric cancers. For example, we sequenced some mammary tumors collected from UC Davis, and one of the dogs with a mammary tumor is only four years old. We also sequenced tumors from older dogs, they are 10 years old or 12 years old. Our sample size is still too small, but by looking for this, the younger dog versus the older dog difference, we can learn. think the carcinogenesis mechanism is different in the younger dogs. I think it's mostly epigenetic change. At four years old, we see really few genomes in terms of the mutation.
We can get a tumor from younger dogs and then compare to the older dogs. It's sort of like comparing pediatric cancers to adult cancer. That's going to be really informative.
Thank you so much, Dr. Zhao. Your lab's key work is in understanding the value of looking at the genomes comparatively, human and dog, the value of big data that looking at thousands upon thousands of cases rather than just a few, how that can bring in tremendous value to genomics. I think almost importantly, just your incredible spirit of collaboration between you and your team, and us at FidoCure® has been absolutely wonderful. And so, I want to leave a few minutes for Christina. I think we have one more question. That is when can we expect this data to be published?
The people actually working on the study are all graduate students that work in my lab collaborating with the FidoCure® data. They are analyzing data and working with FidoCure® scientists closely. Another student's work will compare the protein 3D structures. That's going to be really exciting, because we are working on the tumor specifically and new antigens. They are very devoted with the comparative dog/human work. So I just want to thank them.

In terms of the data, FidoCure® data, this should be published in a couple weeks. It will be the biggest study of canine genomics ever - 700 dogs - the largest one I've ever seen for a single study.
If you would like to get our webinar invites, please send us a note using our veterinarian contact us form.

.png)
